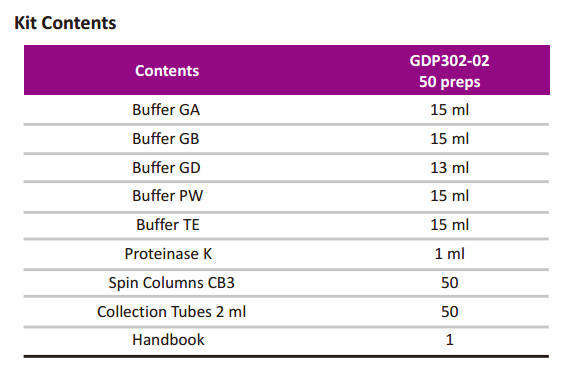
GDP302-TIANamp Bacteria DNA Kit
TIANamp Bacteria DNA Kit—— For isolation of genomic DNA from bacteria
TIANamp Bacteria DNA Kit
(Spin Column)
Cat.no. GDP302
TIANamp Bacteria DNA Kit should be kept in dry place and can be stored
at room temperature (15-30°C) for up to 15 months without showing any
reduction in performance and quality. If any precipitate forms in the buffers,
it should be dissolved by warming the buffers at 37°C for 10 min before use.
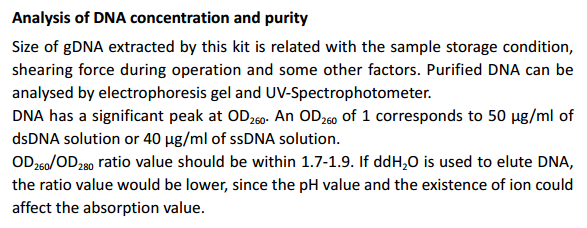
Introduction
TIANamp Bacteria DNA Kit is based on silica membrane technology and
provides special buffer system for many kinds sample of gDNA extraction.
The spin column is made of new type of silica membrane which can bind
DNA optimally on given salt and pH conditions. Simple centrifugation
processing completely removes contaminants and enzyme inhibitors such
as proteins and divalent cations. Purified DNA is eluted in low-salt buffer or
water, and is ready for use in downstream applications.
DNA purified by TIANamp Bacteria DNA Kit is highly suited for restriction
enzyme digestion, PCR analysis, Southern blotting, and cDNA library.
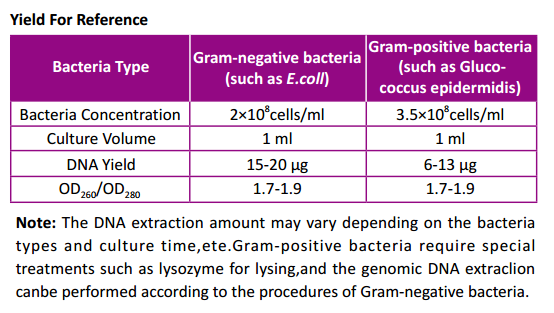
Features
- Simple and fast:Pure genomic DNA of Gram-negative bacteria can be
obtained within 1 hour. - Excellent quality: The purified DNA can be directly used in downstream
molecular experiments such as PCR, restriction endonuclease digestion,
Southern blotting, etc.
Important Notes Please carefully read before using this kit.
- Repeated freezing and thawing of stored samples should be avoided,
since this leads to DNA size reduction. - If precipitates formed in Buffer GA or Buffer GB, warm the buffer to
37°C until the precipitates have fully dissolved. - All centrifugation steps should be carried out in a conventional tabletop centrifuge at room temperature (15-30°C).
Reagents need to be prepared by Customer
96-100% ethanol, RNase A (100 mg/ml) (optional); Lysozyme (50 mg/ml)
(TIANGEN) (optional)
Protocol
Ensure that Buffer GD and Buffer PW have been prepared with appropriate
volume of ethanol (96%-100%) as indicated on the bottle and shake thoroughly.
- Pipet 1-5 ml bacterial culture suspension in a centrifuge tube, centrifuging for 1
min at 10,000 rpm (~11,500 × g), discard supernatant as possible. - Add 200 μl Buffer GA. Mix thoroughly by vortex.
Note: For Gram-positive bacteria, Step 2 could be replaced by lysozyme
treatment: add 110 μl enzymatic lysis buffer (20 mM Tris·Cl, pH 8.0; 2 mM
sodium EDTA; 1.2% Triton® X-100), and 70 μl lysozyme (50 mg/ml prepared
by users). Incubate for at least 30 min at 37°C.
If RNA-Free genomic DNA is required, add 4 μl RNase A (100 mg/ml, should
be prepared by user), mix by vortex for 15 sec, and incubate for 5 min at
room temperature (15-30°C). - Add 20 μl Proteinase K. Mix thoroughly by vortex.
- Add 220 μl Buffer GB to the sample, vortex for 15 sec, and incubate at 70°C for
10 min to yield a homogeneous solution. Briefly centrifuge the 1.5 ml centrifuge
tube to remove drops from the inside of the lid.
Note: White precipitates may form when Buffer GB is added. They will not
interfere with the procedure and will dissolve during the heat incubation
at 70°C. If precipitates do not dissolve during heat incubation, it indicates
that the cell is not completely lysed and may result in low yield of DNA and
impurity in DNA. - Add 220 μl ethanol to the sample, and mix thoroughly by vortex for 15 sec. A
white precipitate may form on addition of ethanol. Briefly centrifuge the 1.5 ml
centrifuge tube to remove drops from the inside of the lid. - Pipet the mixture from step 5 into the Spin Column CB3 (in a 2 ml collection
tube) and centrifuge at 12,000 rpm (~13,400 × g) for 30 sec. Discard flowthrough and place the spin column into the collection tube. - Add 500 μl Buffer GD (Ensure ethanol has been added) to Spin Column CB3,
and centrifuge at 12,000 rpm (~13,400 × g) for 30 sec, then discard the flowthrough and place the spin column into the collection tube. - Add 600 μl Buffer PW (Ensure ethanol has been added) to Spin Column CB3,
and centrifuge at 12,000 rpm (~13,400 × g) for 30 sec. Discard the flowthrough and place the spin column into the collection tube. - Repeat Step 8.
- Centrifuge at 12,000 rpm (~13,400 × g) for 2 min to dry the membrane
completely.
Note: The residual ethanol of buffer PW may affect downstream
application. - Place the Spin Column CB3 in a new clean 1.5 ml centrifuge tube, and pipet
50-200 μl Buffer TE or distilled water directly to the center of the membrane.
Incubate at room temperature for 2-5 min, and then centrifuge for 2 min at
12,000 rpm (~13,400 × g).
Note: If the volume of eluted buffer is less than 50 μl, it may affect
recovery efficiency. The pH value of eluted buffer will have some influence
in eluting. We suggest choosing buffer TE or distilled water (pH 7.0-8.5) to
elute gDNA. For long-term storage of DNA, eluting in Buffer TE and storing
at -30~-15°C is recommended, since DNA stored in water is subject to acid
hydrolysis.
To enhance the gDNA yield, eluate can be reloaded to CB3 once, incubate
at room temperature 2min and centrifuge at 12,000 rpm (~13,400 × g) for 2
min to get the final eluate.
GDP302-TIANamp Bacteria DNA Kit
All trademarks or registered trademarks appearing on this website are the property of their respective owners.
This product is for scientific research use only. Do not use in medicine, clinical treatment, food or cosmetics.
Need more info ? Contact us anytime. We’re here: Go2biotech
E-mail: maggie@go2biotech.com / morgan@go2biotech.com
Telephone:+86 755 8399 5017


