GDP612-DE-TGuide Smart Soil Stool DNA Kit
For purification of inhibitor-free DNA from soil and stool samples.
TECHNICAL MANUAL
Cat. no. GDP612-DE
Note: To use the TGuide Smart Soil/Stool DNA Kit, you must have the
TGuide Smart Soil/Stool DNA (program no. DP612) installed on the TGuide
S16/S32 pro Nucleic Acid Extractor.
Table Contents
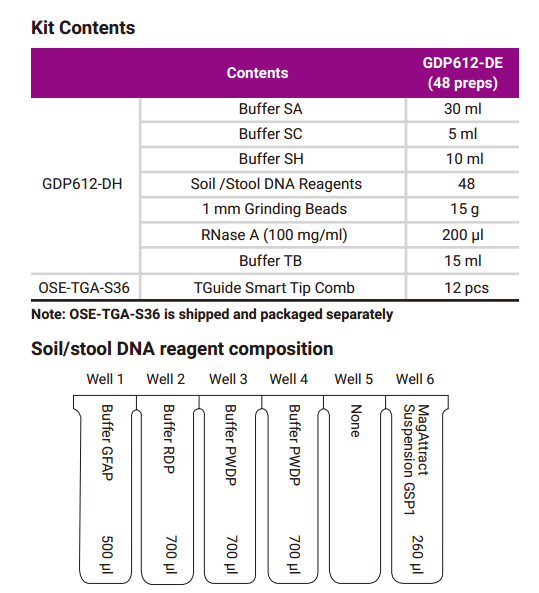
Kit Contents
Soil/Stool DNA reagent composition
Storage condition
Product
Features
Notes
Operational steps
1.Prefilled single sample cartridge
2.Sample pre-processing
3.Operation steps of TGuide S16 Nucleic Acid Extractor
Appendix
1.Program
2.Related Products
TGuide Smart Soil / Stool DNA Kit
Cat. no. GDP612-DE
Storage condition
All components of the kit can be stored in dry conditions at room temperature
(15~30°C) for 12 months. If the solution precipitates, it can be preheated in a
water bath at 37°C for 10 min before use to dissolve the precipitation, without
affecting the effect.
Product
This kit adopts a unique dehumification buffer system to remove humic acid from
soil samples as much as possible. It is also supplied with grinding beads which
effectively break up soil samples in a variety of complex components to ensure
the integrity of genomic DNA extracted from soil. Besides, it is also suitable for
extracting genomic DNA from stool samples.
DNA extracted by this kit has little impurity and good integrity, which can be directly
used for PCR, digestion and other downstream experiments of molecular biology.
Features
- Wide applicability: It is suitable for flower bed soil, flower pot soil, farmland soil,
forest soil, silt, red soil, black soil, dust and other soil environmental samples
extraction, as well as stool sample extraction. - Convenient operation:Ultra-pure genomic DNA can be obtained by running TGuide
S16 for 42 minutes. - High purity:Combined with magnetic bead purification, the extracted DNA with this
kit has high purity and can be directly used in downstream experiments.
Notes
- Fresh samples will ensure a higher yield. For different samples, check the
corresponding optimal storage conditions before sampling. - At the stage where the supernatant needs to be collected, the sediment must be
avoided, otherwise the purity of the product will be affected. - Excessive DNA input may inhibit following PCR reactions. In this case, it is
recommended to dilute the DNA template before use. - Check buffer SC for precipitation before use. If there is precipitation, please heat it
at 37°C until it is completely dissolved before use.
Operational steps
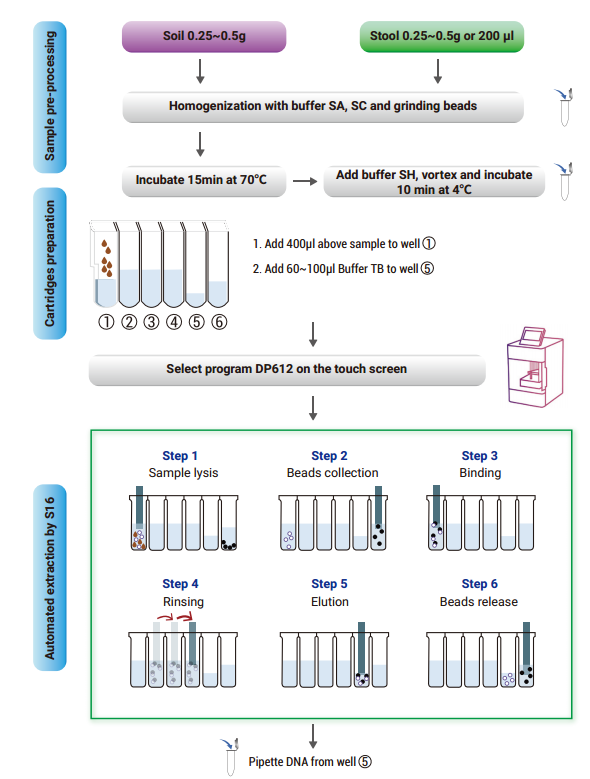
- Prefilled single sample cartridge
1.1 Take out a prefilled single sample cartridge and invert it to re-suspend the
magnetic beads; Gently shake to concentrate the reagent and magnetic beads
to the bottom of the cartridge. Before use, remove sealing film carefully to
avoid liquid spatter or spills.
1.2 Add proper volume (60~100 μl) of elution buffer TB to the 5th well of the
cartridge
- Sample pre-processing
1) Soil sample processing:
Add 0.25~0.5g sample into 2 ml centrifuge tube, as well as 500 μl buffer SA,
100 μl buffer SC and 0.25 g grinding beads for 15 min vortex mixing until the
sample is mixed evenly; or use the TGrinder H24 tissue homogenizer (TIANGEN,
OSE-TH-01, self-prepared) for homogenization (oscillation at 6 M/S speed for 30
sec, with 30 sec interval and 2 cycles). Centrifuge it at 12,000 rpm (~13,400×g)
for 1 min and transfer the supernatant (about 500 μl) to another 2 ml centrifuge
tube.
Note: For some samples with low yield or requirements to extract fungal
genome, it is suggested that after the samples are mixed by vortex mixing or
tissue homogenizer, heat the mixture at 70°C for 15 min to improve the pyrolysis
efficiency.
2) Stool sample processing:
Add 0.25~0.5 g sample into 2 ml centrifuge tube. If the sample is liquid, then
transfer 200 μl sample to the centrifuge tube, and add 500 μl buffer SA, 100
μl buffer SC and 0.25 g grinding beads (Another 4 μl RNase A (100 mg/ml) is
recommended for removing possible residual RNA) for vortex mixing until the
sample is mixed evenly; or use the TGrinder H24 tissue homogenizer (TIANGEN,
OSE-TH-01, self-prepared) for homogenization. Heat the mixture at 70°C for 15
min to improve the pyrolysis efficiency. Centrifuge it at 12,000 rpm (~13,400×g)
for 1 min and transfer the supernatant (about 500 μl) to another 2 ml centrifuge
tube.
Note: For gram-positive bacteria which cell wall are difficult to break , the
temperature can be raised to 95°C to promote the pyrolysis.
2.1 Add 200 μl buffer SH for 5 min vortex mixing and place it at 4 degrees for 10 min.
2.2 Centrifuge it at 12,000 rpm for 2 min and proceed to 3.1.
- Operation steps of TGuide S16 Nucleic Acid Extractor
3.1 Add 400 μl above supernatant to the 1st well of the cartridge and place
cartridges on the reagent tank bracket of TGuide S16 Nucleic Acid Extractor.
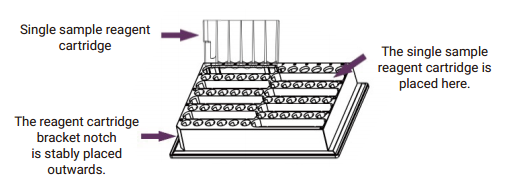
3.2 Place the reagent tank bracket on the plate base in the TGuide S16 Nucleic
Acid Extractor. Insert the Tip Combs into the slot of the Tip Comb to ensure
that they are well connected and firmed
3.3 If you use the TGuide S16 Nucleic Acid Extractor, select the corresponding
program DP612 file on the touch screen, click the icon in the lower right
corner of the screen, or click the “RUN” button at the bottom of the screen to
start the experiment.
3.4 At the end of the automated extraction process, take the DNA out of the 5th
well of the cartridge and store it under appropriate conditions. Single sample
reagent cartridge and tip comb are for single use only.

Appendix
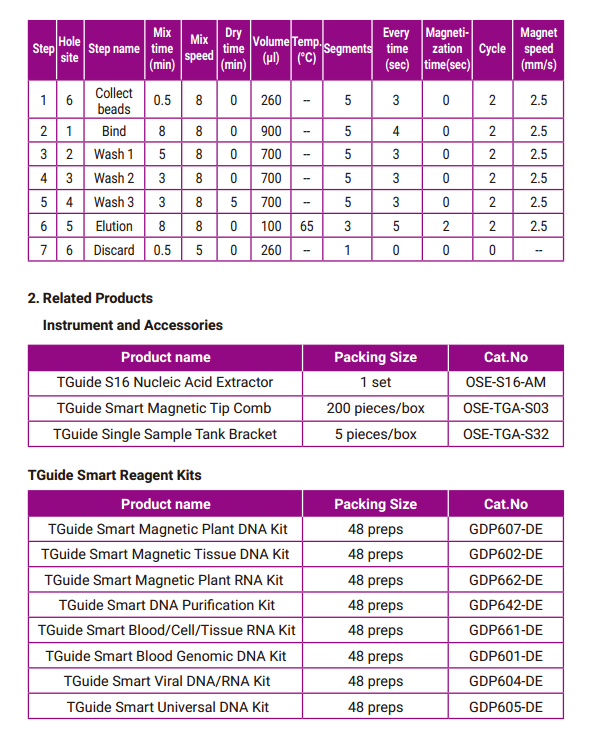
- Program
The extraction process of S16 provided for DP612 is shown in the following table:
GDP612-DE-TGuide Smart Soil Stool DNA Kit
All trademarks or registered trademarks appearing on this website are the property of their respective owners.
This product is for scientific research use only. Do not use in medicine, clinical treatment, food or cosmetics.
Need more info ? Contact us anytime. We’re here: Go2biotech
E-mail: maggie@go2biotech.com / morgan@go2biotech.com
Telephone:+86 755 8399 5017


