NG301-TIANSeq FragmentRepairTailing Module-200528
TIANSeq Fragment/Repair/Tailing Module
Cat.no. 4992350/4992351
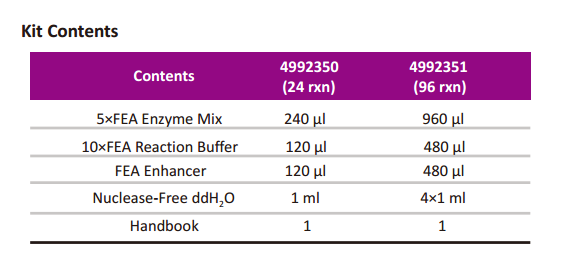
Storage Conditions
TIANSeq Fragment/Repair/Tailing Module could be stored at -30~-15°C for one year. Avoid repeated freezing and thawing.
Product Description
TIANSeq Fragment/Repair/Tailing Module provides a pre-mix module of library
construction for Illumina high-throughput sequencing platforms.
The kit includes all the enzymes for fragmentation, end-repair, and dA-tailing. The
protocol supports fragmentation, end-repair and dA-tailing in a single reaction
step, therefore greatly simplifying the workflow, reducing the total reaction time
and hands-on time. And the product could be used directly for adapter ligation by
TIANSeq Fast Ligation module(Cat# 4992354/4992355).
Application: ideal choice for the DNA fragmentation, end repairing and dA-tailing
for DNA library construction of illumina high-throughput sequencing platform.
DNA input amount: 1 ng-1 µg DNA
Recommended Alternative Reagents
- TIANseq Fast Ligation Module(Cat# 4992354/4992355)
- TIANSeq NGS Library Amplification Module (Cat# 4992373/4992374)
- TIANSeq Single-Indexed Adapter (Illumina)(Cat# 4992641/4992642/4992378)
- TIANSeq Size Selection DNA beads(Cat# 4992358/4992359/4992979)
Product Highlights
- The easily performed one-tube enzymatic reaction can achieve double-strand
DNA fragmentation, end repair and dA-tailing in one step. - High library construction efficiency is achieved with the DNA input as low as 1
ng.
Precautions Please carefully read these precautions before using this kit.
- Attention should be paid in the operating process to avoid cross contamination
between nucleic acid samples and products. - Please use RNase- or DNase-free pipette tips or EP tubes for the experiment.
- Before starting, wipe down work area with RNase and DNase cleaning reagents
such as RNase Away (Molecular BioProducts, Inc). Make sure there is no
contamination of RNase and DNase. - Beforeproceeding related operation, make sure the thermal cycler is calibrated
and in a stable state. - Please read the protocol carefully before the experiment. If test suspension isneeded or the downstream test is not needed to be carried out immediately, thetest products can be frozen and
- Enzyme-based DNA fragmentation is sensitive to many factors, such as reaction
temperature, time, and setup conditions, as well as the input DNA. We strongly
recommend users practicing the protocol and optimizing the parameter
(reaction time) using the same or similar DNA samples.
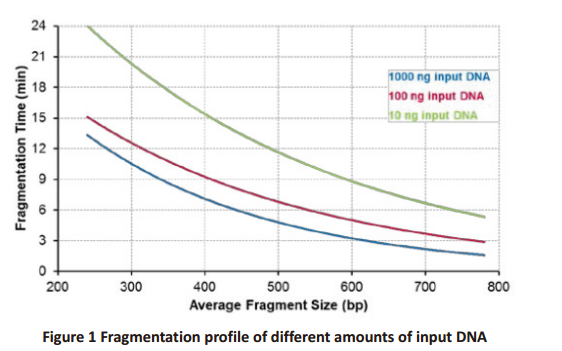
Appendix II Optimization of Fragmentation Time
The reaction time should be optimized for different input DNA (amount and
source). Use the Figure 1 to choose the time that is required to fragmentate
the input DNA to the desired size. The optimization should be carried out using
the same or similar DNA samples that will be used for the final sequencing
experiment. For initial optimization we recommend including 2 additional time
points: 3 minutes longer and 3 minutes shorter than the time calculated from the
figure. Fine tuning may be required if precise fragment size is critical. Optionally,
the fragmentation size can be evaluated immediately after the fragmentation step
if the input DNA amount is ≥100 ng. Use 1.8× TIANSeq Size Selection DNA Beads
(Cat# 4992358/4992359/4992979) to purify the fragmented DNA and elute with
10 μl of Tris buffer or water, then use Bioanalyzer High Sensitivity kit to determine
the size range of the fragmented DNA.
For input DNA <10 ng, to shorten the reaction time, we recommend adding
2.5 μl of Enhancer to the final reaction (50 μl) and use half of the reaction time
determined from the fragmentation profile of 10 ng input DNA in the Table I. For
example, to produce fragment size around 350 bp, after adding the FEA Enhancer,
a 8 minute incubation of the reaction usually generates the expected result.
Appendix III: Fragmentation / End Repair/ dA-tailing of DNA in 1×TE
Follow the instructions below for input DNA dissolved in 1×TE buffer.
- Set up the program as below. Be certain to use the instrument’s heated lid, and
if possible, set the temperature of the heated lid to 70°C.
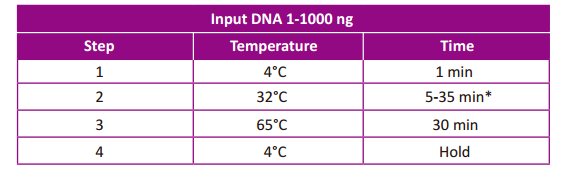
input DNA. For input DNA ≥10 ng, with 2.5 ul FEA Enhancer added in the
reaction, we recommend 25 min as the initial time when it produces
fragment size around 300 to 500 bp. For input DNA <10 ng, with 5 ul FEA
Enhancer added in the reaction, we recommend 15 min as the incubation
time to produce fragment size around 300 bp. Depending on the size
requirement and type of input DNA, either increase or decrease the reaction
time by 3 min incrementally until the expected size range is achieved.
- In order to achieve optimal results, it is important to follow the procedure
described below. Prepare a master mix on ice by combining Fragmentation
Buffer, DNA, and Nuclease-Free ddH2Oas indicated in the table (per DNA
sample).Mix well by gently pipetting (do not vortex).
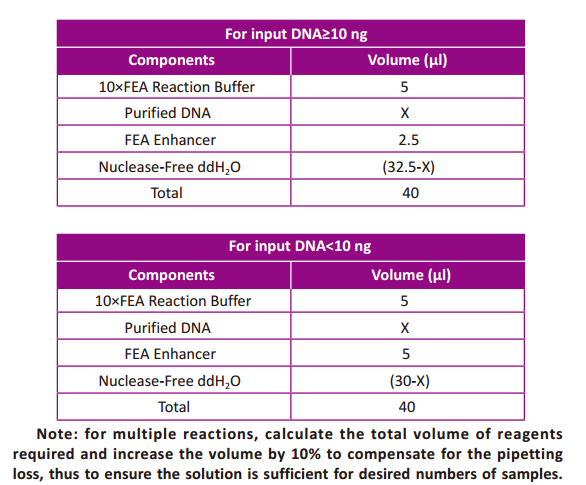
- Transfer 10 μl of 5×FEA Enzyme Mix to a new thin-walled PCR tube for each
reaction. Add 40 μl of the master mix and gently mix well by pipetting up and
down for 10 times. It is critical to keep the PCR tube on ice during the whole
process. - Pulse-spin the sample tube and immediately transfer to the pre-chilled
thermal cycler (4°C). Set up the program. - When thermal cycler program is complete and sample block has returned to
4°C, remove samples from the cycler and place on ice. - Immediately proceed to ligation step. To achieve optimal ligation efficiencies,
we recommend using TIANSeq Fast Ligation Module (Cat# 4992354/ 4992355).
NG301-TIANSeq FragmentRepairTailing Module-200528
All trademarks or registered trademarks appearing on this website are the property of their respective owners.
This product is for scientific research use only. Do not use in medicine, clinical treatment, food or cosmetics.
Need more info ? Contact us anytime. We’re here: Go2biotech
E-mail: maggie@go2biotech.com / morgan@go2biotech.com
Telephone:+86 755 8399 5017


